pacman::p_load(tidyverse,sf,funModeling)In-class Exercise 2: Geospatial Data Wrangling
Load Packages and Data
sf: handle geospatial data
tidyverse: for data science work (data handling, manual processing and manipulation)
funModeling: quick exploratory data analysis
Geoboundaries dataset
Transform data’s WGS84 format to the Projected Coordinate Systems of Nigeria’s. All data will now be in metres.
geoNGA <- st_read("data/geospatial", layer = "geoBoundaries-NGA-ADM2")%>%
st_transform(crs = 26392)Reading layer `geoBoundaries-NGA-ADM2' from data source
`C:\zoe-chia\IS415\In-class_Ex\In-class_Ex02\data\geospatial'
using driver `ESRI Shapefile'
Simple feature collection with 774 features and 6 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 2.668534 ymin: 4.273007 xmax: 14.67882 ymax: 13.89442
Geodetic CRS: WGS 84NGA dataset
NGA <- st_read("data/geospatial", layer = "nga_admbnda_adm2_osgof_20190417")%>%
st_transform(crs = 26392)Reading layer `nga_admbnda_adm2_osgof_20190417' from data source
`C:\zoe-chia\IS415\In-class_Ex\In-class_Ex02\data\geospatial'
using driver `ESRI Shapefile'
Simple feature collection with 774 features and 16 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 2.668534 ymin: 4.273007 xmax: 14.67882 ymax: 13.89442
Geodetic CRS: WGS 84Aspatial data
Waterpoint dataset
Dataset consists of the entire world’s waterpoints. Hence, filter and get only those with “Nigeria” mentioned.
wp_nga <- read_csv("data/aspatial/WPdx.csv") %>%
filter(`#clean_country_name` == "Nigeria")Converting water point data into sf point features
Aspatial data does not have spatial properties. Hence, to turn it into geospatial data, we need to (i) convert it into the appropriate data type and (ii) add projection information (iii) transform to the respective coordinate system:
- Convert the wkt field into sfc field by using st_as_sfc() data type.
wp_nga$Geometry = st_as_sfc(wp_nga$`New Georeferenced Column`)
wp_nga# A tibble: 95,008 × 71
row_id `#source` #lat_…¹ #lon_…² #repo…³ #stat…⁴ #wate…⁵ #wate…⁶ #wate…⁷
<dbl> <chr> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr>
1 429068 GRID3 7.98 5.12 08/29/… Unknown <NA> <NA> Tapsta…
2 222071 Federal Minis… 6.96 3.60 08/16/… Yes Boreho… Well Mechan…
3 160612 WaterAid 6.49 7.93 12/04/… Yes Boreho… Well Hand P…
4 160669 WaterAid 6.73 7.65 12/04/… Yes Boreho… Well <NA>
5 160642 WaterAid 6.78 7.66 12/04/… Yes Boreho… Well Hand P…
6 160628 WaterAid 6.96 7.78 12/04/… Yes Boreho… Well Hand P…
7 160632 WaterAid 7.02 7.84 12/04/… Yes Boreho… Well Hand P…
8 642747 Living Water … 7.33 8.98 10/03/… Yes Boreho… Well Mechan…
9 642456 Living Water … 7.17 9.11 10/03/… Yes Boreho… Well Hand P…
10 641347 Living Water … 7.20 9.22 03/28/… Yes Boreho… Well Hand P…
# … with 94,998 more rows, 62 more variables: `#water_tech_category` <chr>,
# `#facility_type` <chr>, `#clean_country_name` <chr>, `#clean_adm1` <chr>,
# `#clean_adm2` <chr>, `#clean_adm3` <chr>, `#clean_adm4` <chr>,
# `#install_year` <dbl>, `#installer` <chr>, `#rehab_year` <lgl>,
# `#rehabilitator` <lgl>, `#management_clean` <chr>, `#status_clean` <chr>,
# `#pay` <chr>, `#fecal_coliform_presence` <chr>,
# `#fecal_coliform_value` <dbl>, `#subjective_quality` <chr>, …- Convert tibble dataframe into an sf object using st_sf(). It is also important for us to include the referencing system of the data into the sf object. Data in wp_nga is in decimals, and in tibble form. It does not have the projection information. We need to put back its original projection inform (WGS84).
wp_sf <- st_sf(wp_nga, crs = 4326)
wp_sfSimple feature collection with 95008 features and 70 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: 2.707441 ymin: 4.301812 xmax: 14.21828 ymax: 13.86568
Geodetic CRS: WGS 84
# A tibble: 95,008 × 71
row_id `#source` #lat_…¹ #lon_…² #repo…³ #stat…⁴ #wate…⁵ #wate…⁶ #wate…⁷
* <dbl> <chr> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr>
1 429068 GRID3 7.98 5.12 08/29/… Unknown <NA> <NA> Tapsta…
2 222071 Federal Minis… 6.96 3.60 08/16/… Yes Boreho… Well Mechan…
3 160612 WaterAid 6.49 7.93 12/04/… Yes Boreho… Well Hand P…
4 160669 WaterAid 6.73 7.65 12/04/… Yes Boreho… Well <NA>
5 160642 WaterAid 6.78 7.66 12/04/… Yes Boreho… Well Hand P…
6 160628 WaterAid 6.96 7.78 12/04/… Yes Boreho… Well Hand P…
7 160632 WaterAid 7.02 7.84 12/04/… Yes Boreho… Well Hand P…
8 642747 Living Water … 7.33 8.98 10/03/… Yes Boreho… Well Mechan…
9 642456 Living Water … 7.17 9.11 10/03/… Yes Boreho… Well Hand P…
10 641347 Living Water … 7.20 9.22 03/28/… Yes Boreho… Well Hand P…
# … with 94,998 more rows, 62 more variables: `#water_tech_category` <chr>,
# `#facility_type` <chr>, `#clean_country_name` <chr>, `#clean_adm1` <chr>,
# `#clean_adm2` <chr>, `#clean_adm3` <chr>, `#clean_adm4` <chr>,
# `#install_year` <dbl>, `#installer` <chr>, `#rehab_year` <lgl>,
# `#rehabilitator` <lgl>, `#management_clean` <chr>, `#status_clean` <chr>,
# `#pay` <chr>, `#fecal_coliform_presence` <chr>,
# `#fecal_coliform_value` <dbl>, `#subjective_quality` <chr>, …- Transform to Nigeria’s projected coordinate system
wp_sf <- wp_sf %>%
st_transform(crs = 26392)Geospatial Data Cleaning
Excluding redundant fields
filter() - selects rows that you want to retain
select() - selects columns that you want to retain
NGA <- NGA %>%
select(c(3:4, 8:9))Checking for duplicate names
Using duplicated() of Base R, we can flag out LGA names that might be duplicated as shown in the code chunk below.
NGA$ADM2_EN[duplicated(NGA$ADM2_EN)==TRUE][1] "Bassa" "Ifelodun" "Irepodun" "Nasarawa" "Obi" "Surulere"The above output shows us that those are the duplicated records.
To fix this, add the state (ADM1_EN) behind.
NGA$ADM2_EN[94] <- "Bassa, Kogi"
NGA$ADM2_EN[95] <- "Bassa, Plateau"
NGA$ADM2_EN[304] <- "Ifelodun, Kwara"
NGA$ADM2_EN[305] <- "Ifelodun, Osun"
NGA$ADM2_EN[355] <- "Irepodun, Kwara"
NGA$ADM2_EN[356] <- "Irepodun, Osun"
NGA$ADM2_EN[519] <- "Nasawara, Kano"
NGA$ADM2_EN[520] <- "Nasawara, Nasawara"
NGA$ADM2_EN[546] <- "Obi, Benue"
NGA$ADM2_EN[547] <- "Obi, Nasawara"
NGA$ADM2_EN[693] <- "Surulere, Lagos"
NGA$ADM2_EN[694] <- "Surulere, Oyo"Check that duplicates have been handled:
NGA$ADM2_EN[duplicated(NGA$ADM2_EN)==TRUE]character(0)Data Wrangling for Water Point Data
freq() of funModeling package is used to reveal the distribution of water point status visually.
freq(data = wp_sf,
input = '#status_clean')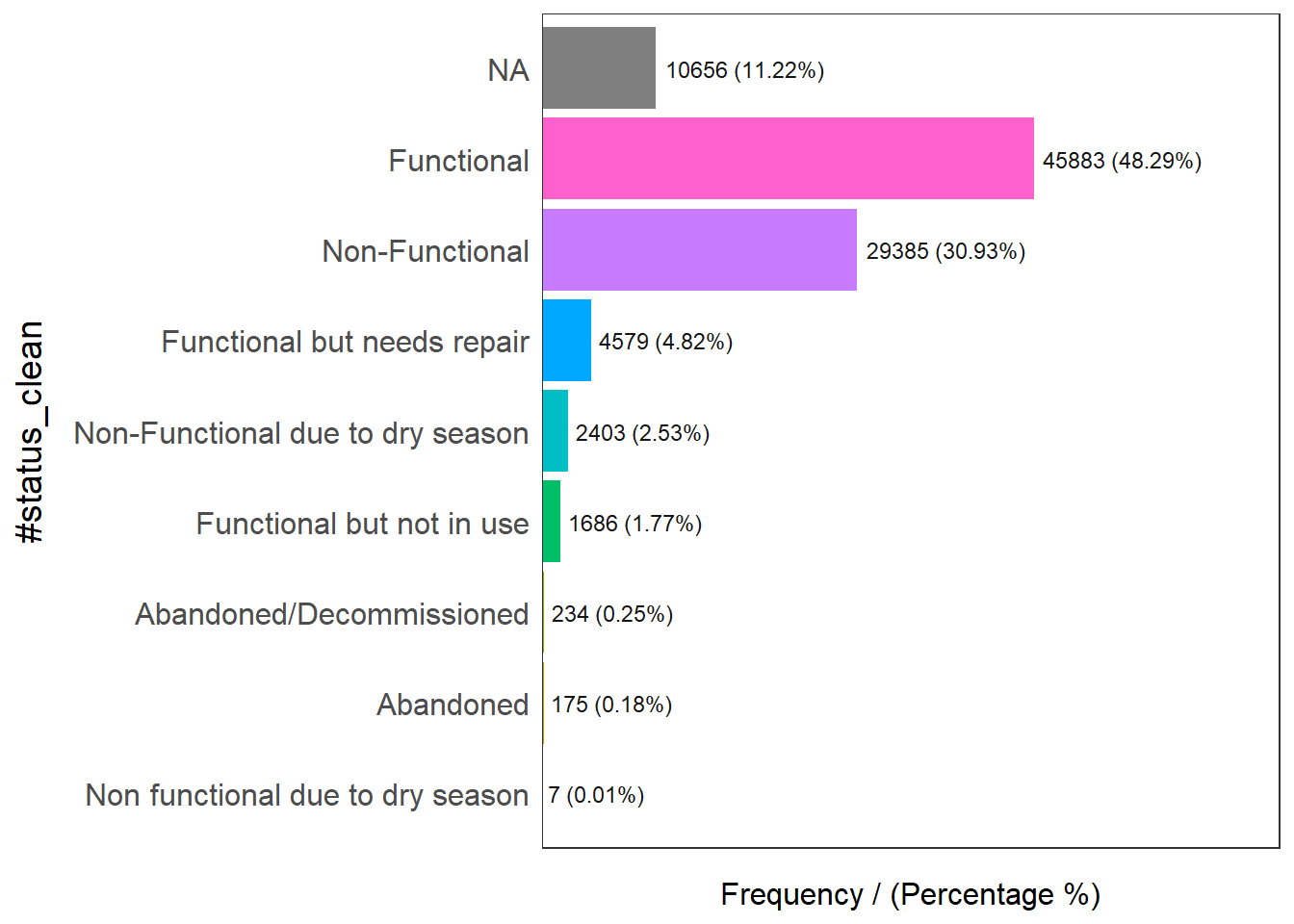
#status_clean frequency percentage cumulative_perc
1 Functional 45883 48.29 48.29
2 Non-Functional 29385 30.93 79.22
3 <NA> 10656 11.22 90.44
4 Functional but needs repair 4579 4.82 95.26
5 Non-Functional due to dry season 2403 2.53 97.79
6 Functional but not in use 1686 1.77 99.56
7 Abandoned/Decommissioned 234 0.25 99.81
8 Abandoned 175 0.18 99.99
9 Non functional due to dry season 7 0.01 100.00Rename columns
Rename NA so we can do mathematical calculations.
rename() - Remove “#”, for example from ‘#status_clean’ to status_clean for easier handling in subsequent steps.
select () - To select the column status_clean in the output of the sf dataframe.
mutate() & replace_na() to recode all the NA values in status_clean into ‘unknown’
wp_sf_nga <- wp_sf %>%
rename(status_clean = '#status_clean') %>%
select(status_clean) %>%
mutate(status_clean = replace_na(
status_clean, "unknown"
))Extracting Water Point Data
The code chunk below is used to extract functional water points.
wp_functional <- wp_sf_nga %>%
filter(status_clean %in%
c("Functional",
"Functional but not in use",
"Functional but needs repair"))Extract non-functional water points:
wp_nonfunctional <- wp_sf_nga %>%
filter(status_clean %in%
c("Abandoned/Decommissioned",
"Abandoned",
"Non-Functional due to dry season",
"Non-Functional",
"Non functional due to dry season"))Extract water points with unknown status:
wp_unknown <- wp_sf_nga %>%
filter(status_clean == "unknown")Point-in-Polygon Count
Identify functional water points in each LGA by using st_intersects() of the sf_package.
Next, use lengths() to calculate the number of functional water points that fall inside each LGA.
NGA_wp <- NGA %>%
mutate(`total_wp` = lengths(
st_intersects(NGA, wp_sf_nga))) %>%
mutate(`wp_functional` = lengths(
st_intersects(NGA, wp_functional))) %>%
mutate(`wp_nonfunctional` = lengths(
st_intersects(NGA, wp_nonfunctional))) %>%
mutate(`wp_unknown` = lengths(
st_intersects(NGA, wp_unknown)))These 4 new fields (total_wp, wp_functional, wp_nonfunctional, wp_unknown) are created in NGA_wp.
Visualing attributes by using statistical graphs
gglplot2 package is used to reveal the distribution of total water points by LGA in a histrogram.
ggplot(data = NGA_wp,
aes(x = total_wp)) +
geom_histogram(bins=20,
color="black",
fill="light blue") +
geom_vline(aes(xintercept=mean(
total_wp, na.rm=T)),
color="red",
linetype="dashed",
size=0.8) +
ggtitle("Distribution of total water points by LGA") +
xlab("No. of water points") +
ylab("No. of\nLGAs") +
theme(axis.title.y=element_text(angle = 0))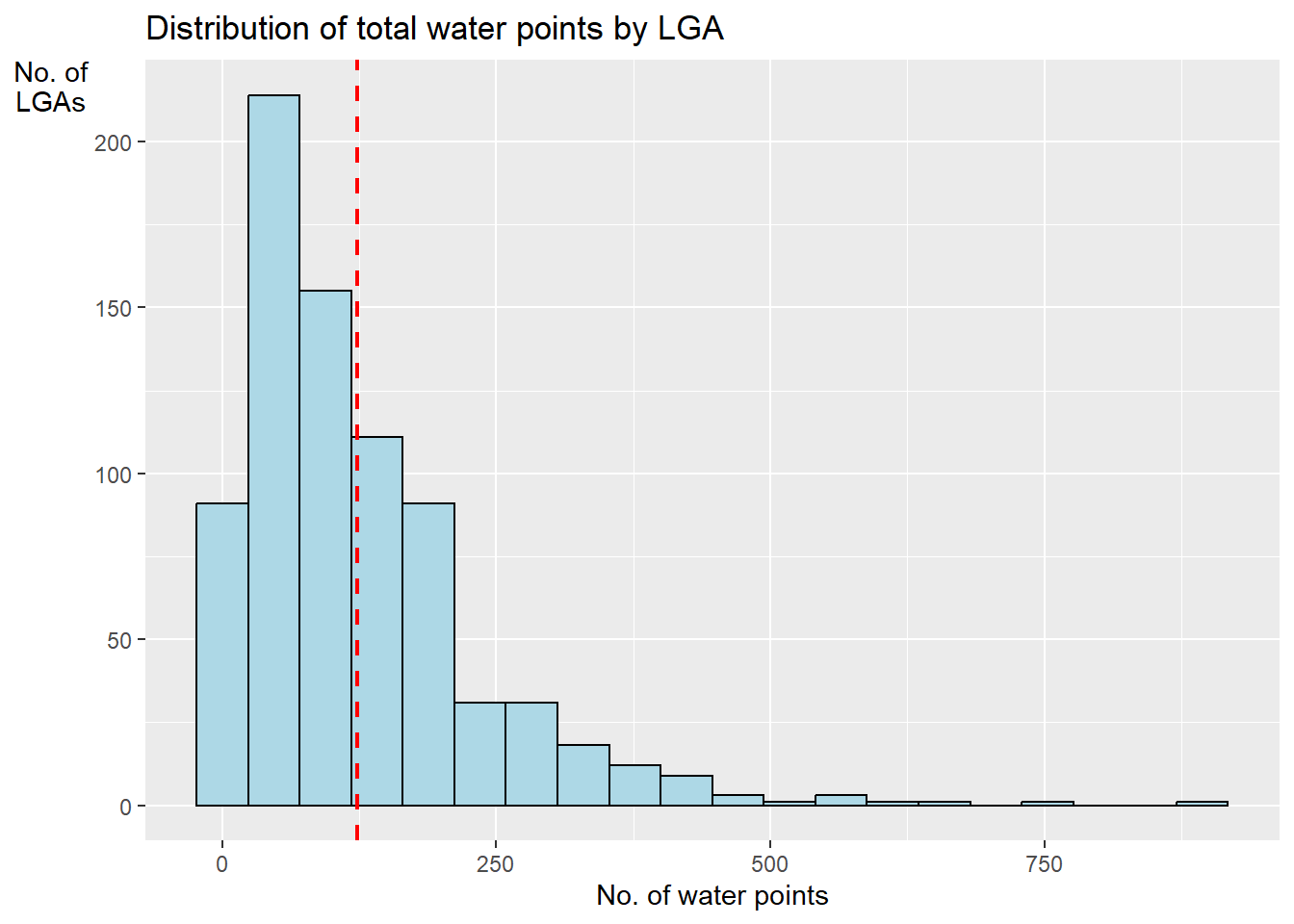
Save the analytical data in rds format
It is recommended retain the sf object structure for subsequent analysis by saving the sf data.frame into rds format.
write_rds(NGA_wp, "data/rds/NGA_wp.rds")